Next: Sparseness and sampling zeros Up: Collapsibility Previous: Collapsibility
genlog freq size mode
/print=est/plot=none
/design = freq size mode mode*size
freq*mode freq*size.
The two estimated parameters for the freq*size
interaction are 0.5828 and 0.3895, for the effect of
large and medium respectively, versus small, the reference
category. Thus the difference between large and medium is
approximately half the difference between medium and small. What
happens if we redefine size as large/medium versus small?
recode size (1,2=1) (3=2) into size2.
genlog freq size2 mode
/print=est/plot=none
/design = freq size2 mode mode*size2
freq*mode freq*size2.
Deviance falls from 4.1415 for 4 df to 0.2633 for 2 df. They are
both very well fitting models, and the difference between them is
slight (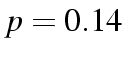 ), but the simpler model has the virtue of being
simpler to interpret.
), but the simpler model has the virtue of being
simpler to interpret.
 |
| ||||||||