Exploit your cores!
If you don’t have Stata-MP, it can be difficult to benefit from all the cores on your computer. However, if your problem can be split up in parts that can run in parallel, it is easy to run multiple instances of Stata. In this note I demonstrate a simple case, using the example of a simulation I wish to run many times.
To do this, you need to write your program to take parameters so that it can be invoked multiple times but produce different results. To avoid having all instances writing to the same log file, the source do-file needs to be renamed or placed in multiple locations. In this example I create subdirectories and place copies there, but you could simply copy the source do-file to numbered copies.
The command-line parameters can include things like a random seed, or other settings that need to differ across runs.
I present a pretty simple example simulation: given a true ranking that is based on a true continuous score variable, what does measurement error in the ranking look like if there is measurement in the score variable? I simulate 100 cases, calculating their true rank from the true score (a standard normal random variable), and then a second rank based on the score with a certain amount of random-normal error added (a normal random variable, mean zero, standard deviation given by a parameter). My summary statistic for each replication is the mean across the 100 cases of the absolute difference between the ranks; I store this in a .dta file using Stata’s postfile framework. For each value of the error parameter (a list between 0.01 and 1), I want, say, 10,000 replications. On my desktop, I have 8 cores, so we can run 8 times 1,250 replications at once.
To set the 8 instances running, I have written a little unix shell-script. Stata could also be used to carry out this task, or a DOS .bat file.
for a in {1..8}; do
mkdir -p run$a
cp -p ranktest_mp.do run${a}/
( cd run$a && stata -b do ranktest_mp "$RANDOM 1250" ) &
done
This creates the 8 required subdirectories (mkdir -p will not trigger an error if they already exist), and copies the ranktest_mp.do file to each one. The commands in parentheses change directory to each subdirectory and invoke Stata on the file, passing as a single string two parameters, one the random seed (using the Bash built-in RANDOM environment variable) and the number of replications each instance is to do. The “&” at the end makes these run in the background, and means the eight runs will be triggered in parallel.
The code in the do-file begins by accessing the command line parameters, setting up the postfile, and setting (and displaying, for the record) the random seed
// Take arguments from the command line args seed replications // Open a file to post the results to postfile pf error meandiff using ranktest, every(10) set seed `seed' di "Seed: `c(seed)'"
We then have a pair of loops, the outer one repeating as many times as the replications parameter, and the inner one repeating the calculation for a number of levels of measurement error. At the end of each inner loop, the results (both the error parameter and the summary statistic) are posted to the postfile. When the outer loop terminates, the postfile is closed.
// for each error level, run many simulations
forvalues x = 1/`replications' {
foreach error of numlist 0.01 0.05 0.1 0.2 0.5 1.0 {
clear
set obs 100
gen score = rnormal()
sort score
gen rank = _n
gen score2 = score + rnormal(0,`error')
sort score2
gen rank2 = _n
gen absdiff = abs(rank - rank2)
qui su absdiff
post pf (`error') (`r(mean)')
}
}
postclose pf
At the end we need to gather the results and report a summary. We have 8 subdirectories each containing a file =ranktest.dta=. This is easily done:
use run1/ranktest
gen run = 1
forvalues x = 2/8 {
append using run`x'/ranktest
replace run = `x' if missing(run)
}
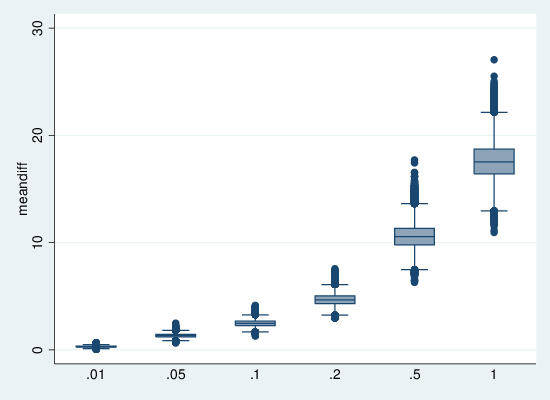
graph box meandiff, over(error)
Timing
You can play around with using different numbers of cores. In my experience the more cores you use the less the additional improvement (so for instance, 8 cores are not twice as fast as 4) but still the more cores used the less time taken overall. One way to capture the time info on Unix is to wrap the Stata invocation in the time command:
for a in {1..8}; do
mkdir -p run$a
cp -p ranktest_mp.do run${a}/
( cd run$a && time stata -b do ranktest_mp "$RANDOM 1250" ) &
done
Alternatively use Stata’s timer command.
Summary
When you have a simply parallelisable problem (like a simulation) it’s easy to split and run it over multiple cores. The key trick is to make multiple copies of the do-file and to pass parameters to it.
Hi Brendan,
This example has been very helpful for me. I am running a simulation on Stata and would like to use all cores (the model fits I am using have not been parallelized in Stata/MP).
You have written the little unix shell-script and say Stata could also be used to carry out this task, or a DOS .bat file. Do you have an example of how this code could be written Stata or a DOS .bat file to run the Stata instances in parallel? I am struggling to translate the unix shell script as a non-unix user.
Jane
Hi Jane,
The main code translated to Stata might look something like this, in part:
forvalues x = 1/8 {
mkdir run`x’
copy ranktest_mp.do run`x’/
cd run`x’
winexec stata -b do ranktest_mp “`=runiformint(0,99999)’ 1250”
cd ..
}
I don’t have access to Stata for Windows at the moment, so I can’t test it, and I am not 100% sure winexec will behave.