Speeding up embarrassingly parallel work in Stata
Stata-MP enables parallel processing, but this really kicks in only in estimation commands that are programmed to exploit it. A lot of the time we may have very simply repetitive tasks for Stata to do, that could be packaged for parallel operation very easily. This sort of work is often called embarrassingly parallel, and isn’t automatically catered for by Stata-MP.
The simplest situation is where the same work needs to be done multiple times, with some different details each time but no dependence whatsoever on the other parts. For instance we may have a loop where the work in the body takes a long time but doesn’t depend on the results of the other runs.
I came across this situation the other day: I have data created by running a simulation with different values of two key parameters, and I wanted to create graphs for each combination of the parameters (PNGs rather than Stata graphs, but that’s just a detail). So I wrote code like the following:
forvalues i = 0/20 {
forvalues j = 1/8 {
[... complicated commands making the graph, depending in `i' and `j' ...]
}
}
OK, so that specified 21 times 8 = 168 graphs. But each one took about 25 seconds to generate, so it takes over an hour. On the other hand, it uses only one core, and my computer has eight. Eight? What if I ran this code eight times, varying the 1 in the second line:
use simdata local j 1 forvalues i = 0/20 { [... complicated commands making the graph, depending in `i' and `j' ...] }
In fact, if the code looks like this and resides in smallfile.do:…
use simdata local j `1' forvalues i = 0/20 { [...] }
…I can run it from within Stata as follows (varying the 1 from 1 to 8):
!stata -b do smallfile.do 1 &
Under Unix, the “&” at the end makes this work in the background, as long as the “!stata -b do …” command is issued in an interactive Stata session.
There is a slight problem: I can’t run files with the same name simultaneously in the same folder, without Stata fighting with itself about writing to the log files. One strategy I’ve used before is to make each command work in its own subfolder: that’s good but requires a bit more prep. What I did yesterday was to copy the smallfile.do to temporary files:
forvalues x = 1/8 {
tempname aux`x'
!cp -p smallfile.do `aux`x''.do
!stata -b do `aux`x''.do `x' &
}
This creates files named like __000001.do, and Stata logs to files like __000001.log. Since the outputs (PNG-files) are named depending on the `x' value, they don’t clash.
Finally, I realised that the “!” or shell command in Stata will run in the background (asychronously, in parallel) only if it is invoked in an interactive Stata session. If the main file is run in batch mode, the sub-tasks are executed sequentially. So I tried the winexec command, which according to the documentation is intended explicitly for calling graphical programs like a web browser, while in interactive sessions. But it works:
forvalues x = 1/8 {
tempname aux`x'
!cp -p smallfile.do `aux`x''.do
winexec stata -b do `aux`x''.do `x'
}
winexec, without an &, will run the subtasks in parallel, even from batch mode!
Summary
This strategy depends on the subtasks being independent: while they all use the same .dta file, they don’t depend on each other, and they produce output that is named independently and doesn’t clash. The smallfile.do takes a command line parameter, so it operates differently for each run. It uses this parameter to name the outputs (e.g., gr_`x'_`1'.png). Copying the smallfile.do to a temporary file (or functionally equivalently, to a run-specific folder) means that the parallel Stata tasks don’t clash with each other. The temporary files should be handled more cleanly (check they don’t exist first, delete them after use, etc), but are easy to remove afterwards.
This example creates lots of graphs. We could equally generate new data files, and use a simple append to pull them together afterwards:
clear forvalues x = 1/8 { append using smalldata`x' }
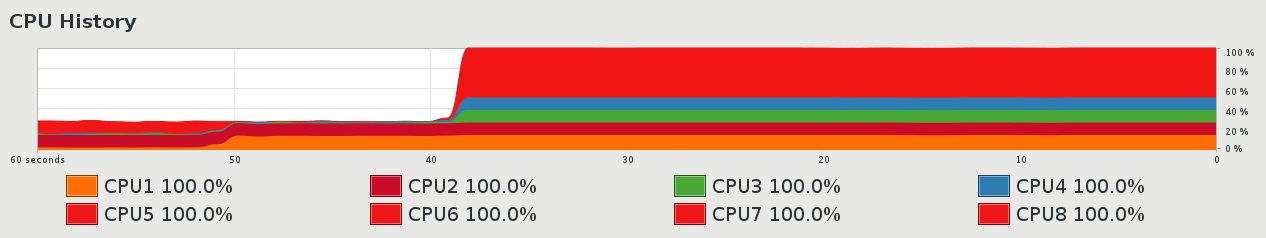
It’s a good speedup: I get my PNG files generated at 16/min rather than 2.5/min, so it gets done in about 10 minutes rather than about an hour. Easy to code and saves time. And it makes my CPU monitor look like this: